
Cannot recreate trace with polynomial fit coefficients
I am a bit entangled at the moment. Maybe it is something easy that I am missing...
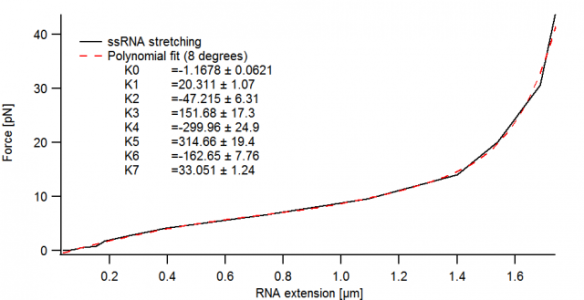
I have a wave, to which I fitted a polynome of 8 degrees. Now, when I want to recreate the trace with the fitting coefficients only, I get something completely different and drastically off from the values and range I expected.
Please find attached first the fit with the coefficients (Fig1) and secondly the resulting recreated trace (Fig2).
What I did is the following:
MAKE/O/N=(427) ssRNAForce, ssRNAextension // making new waves, ranging from 0 to 42.6 pN, with 0.1 pN increment
WAVE ssRNAForce, ssRNAextension
ssRNAForce =x*0.1
VARIABLE K0 =-1.1678 // coefficients of previous polynomial fit (8 degrees)
VARIABLE K1 =20.311
VARIABLE K2 =-47.215
VARIABLE K3 =151.68
VARIABLE K4 =-299.96
VARIABLE K5 =314.66
VARIABLE K6 =-162.65
VARIABLE K7 =33.051
ssRNAextension = K0 + (K1 *ssRNAForce) + (K2 * ssRNAForce^2) + (K3*ssRNAForce^3) + (K4*ssRNAForce^4) + (K5*ssRNAForce^5) + (K6*ssRNAForce^6) + (K7*ssRNAForce^7) // recreate the trace
I'd be very happy if someone could point me to the right direction.
Many thanks.







Look at the values in the coefficients wave after you have performed the fit.
Hope this helps,
Kurt
October 28, 2016 at 02:41 am - Permalink
Thanks for the prompt reply and idea.
I placed now values with 16 digits, but the results is unfortunately the same.
Any other ideas?
Regards.
October 28, 2016 at 03:20 am - Permalink
Additionally you only have a few decimal places for the K variables in your code.
October 28, 2016 at 04:09 am - Permalink
Also thanks to your idea of the /D flag.
Based on Kurt's comment, I already took into account 16 digits. I tried it also by just using directly the values of the W_coef wave.
However, nothing changed.
Code is currently:
MAKE/O/D/N=(427) ssRNAForce, ssRNAextension
WAVE ssRNAForce, ssRNAextension
ssRNAForce =x*0.1
VARIABLE K0 = -1.167821346598224
VARIABLE K1 =20.31138282343296
VARIABLE K2 =-47.21531523237702
VARIABLE K3 =151.6787962105859
VARIABLE K4 =-299.9575742262949
VARIABLE K5 =314.6620434878205
VARIABLE K6 =-162.6499098196241
VARIABLE K7 =33.05068128748616
ssRNAextension = K0 + (K1 *ssRNAForce) + (K2 * ssRNAForce^2) + (K3*ssRNAForce^3) + (K4*ssRNAForce^4) + (K5*ssRNAForce^5) + (K6*ssRNAForce^6) + (K7*ssRNAForce^7)
I have still no idea...
October 28, 2016 at 04:17 am - Permalink
With the following modification to your function; I get a plot similar to your initial data. Note also the change to the assignment of values to ssRNAextension.
October 28, 2016 at 05:44 am - Permalink
It is akin to the parable of the blind men describing the elephant. With seven blind men, you get a description of the elephant. With eight, you get its tail to wag too.
I might go a step further and hazard a suggestion. If this is data for the stretching force of a bond as a function of distance r, a better choice might be to use a parametric model. For example, a Hook's law expression FHL = kr or a "Lennard-Jones" expression FLJ = C/r^n.
--
J. J. Weimer
Chemistry / Chemical & Materials Engineering, UAH
October 28, 2016 at 07:04 am - Permalink
The next step would be to fix the power law exponent to an integer. It seems to be 12 or 13.
--
J. J. Weimer
Chemistry / Chemical & Materials Engineering, UAH
October 28, 2016 at 07:42 am - Permalink
2) In Fig2.png, the range of the axis goes out to 7e12. Try computing 7e12^7 and then add it to a constant term -1.1678. Even double-precision can't do that.
I go with the explanation of confused input waves in the computation.
And JJWeimer, nice job!
John Weeks
WaveMetrics, Inc.
support@wavemetrics.com
October 28, 2016 at 09:35 am - Permalink
@ jtigor: Yeah, it works. It is however a bit strange to me, as it should be the right way; at least it does always for my other stuff this way around. Anyway, this simple change did the trick. Thanks!
@ J.J.: You are absolutely right. One should look to the physical meaning and decide for a model which fits. In principle, here we have an elastic biopolymer. The elasticity of biopolymers, probed using single molecule methods, is often analyzed using the worm-like chain (WLC) model or the freely-jointed chain (FJC) model. The WLC works for dsDNA and ssRNA likewise, whereas, in contrast, for ssDNA the FJC model works better. There is always a bit of controversy discussion in the literature. However, I don't want to go to deep to this topic now; in fact, we have here a mixture of single and double strand RNA (fraction of ssRNA is ~80%). So, we cannot fit easily with a standard model. One could go further trying to find/modify a model, but this curve should only be a reference of empirical measured elasticity. We don't go further into any polymer physics here as we need just boundaries.
Your example, however, to combine two fits is actually neat. It doesn't work for this curve (see attached Fig3 with the measured data at log scale), as it does not behave linearly at lower forces. But I could use this approach for another curve. Here, the polynomial reconstruction is the only one that satisfied me sufficiently to be able to work with it as boundary. The main reason for such simple (and non-physical) reconstruction is that I don't want to load the data every time a user just wants to now the average extension at a specific force.
If you want to know in more detail (as you like the force topic) for what this actually is, you could take a look to this paper including supplemental info: http://dx.doi.org/10.1016/j.celrep.2015.01.03
@ John: Thanks for your reply. Yes, I know that it is in real a number of 7 terms, but I wanted to make clear what I performed in Igor. I could have described that a bit more in detail, indeed. However, I did not understand your second point in terms of ‘computing 7e12^7’. I’m a biologist ;-)
October 28, 2016 at 09:47 am - Permalink
A composite? Based on fraction? Neat!
The doi link fails. Can you correct this?
Oh. Oh. Oh! I wonder if this would work with F = k*r + C1/r^n1 -C2/r^n2. That is still down from 8 coefficients to 5.
But, I'm getting a bit too much in to my hobby (curve fitting) when I have other real-work stuff to do.
--
J. J. Weimer
Chemistry / Chemical & Materials Engineering, UAH
October 28, 2016 at 11:03 am - Permalink
Oh. Sorry, for the inconvenience that the link doesn’t work properly.
You may directly download the PDF that includes also the SI here:
http://nynkedekkerlab.tudelft.nl/wp-content/uploads/dekker_cell_reports…
For the moment, I am fine with the polynomial fit. But thanks for the idea.
If forces and curve fitting is your hobby, I may then get to you when I have some interesting puzzles to solve ;-)
Cheers! :-)
October 28, 2016 at 12:31 pm - Permalink
October 28, 2016 at 04:52 pm - Permalink
Puzzles in fitting force curves is actually one of my new adventures. I'd be glad to hear more as you find the interest.
--
J. J. Weimer
Chemistry / Chemical & Materials Engineering, UAH
October 28, 2016 at 01:51 pm - Permalink
Being a biologist is something that can be overcome!
Anyway, I see 7e12 as the tick mark label on the right end of Fig2.png. So the polynomial with 8 terms will have X^7, or 7e12^7 = 8.23543e+89. Since a double-precision number has about 16 digits of precision, adding the constant term (-1.1678) to that will make absolutely no difference. In fact, the first several terms will be completely washed out, no doubt. And the alternating signs will result in subtractions that are small differences of large numbers, with loss of floating-point accuracy.
John Weeks
WaveMetrics, Inc.
support@wavemetrics.com
October 28, 2016 at 02:37 pm - Permalink
Absolutely, I am working on it with excitement.
Thanks for clarifying. Now, I understood what you meant.
Cheers.
October 28, 2016 at 04:58 pm - Permalink