
NeuroTools: an interface for flexible data organization and analysis
Background
NeuroTools provides an interface for efficiently navigating, organizing, and performing analysis routines on large data sets.
NeuroTools was originally written with electrophysiological and functional imaging data in mind, but is now developed as a general purpose analysis suite. With the ability to add user analysis packages, NeuroTools can be as expandable as the user wants. Many of the data organization concepts in NeuroTools were inspired by previous work by Dr. W. Rowland Taylor at Berkeley and Dr. Rob Smith at Penn.
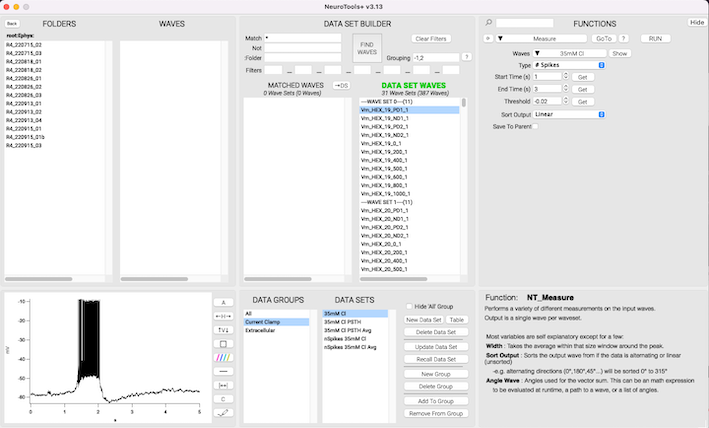
At its core, NeuroTools is an alternative data browser that allows users to define, save, and recall 'Data Sets' using powerful wave matching tools. Data Sets can be organized and filtered in a variety of ways, allowing the user to customize how they run batches of waves through a function.
Functions and Packages
While NeuroTools provides a suite of basic built-in functions, users can easily write their own add-on package, or import any of the specialized packages available from https://www.benmurphybaum.com/NeuroToolsStore.html
Below is an example of selecting a function from the ScanImage package, which is a downloadable add-on designed to load, explore, and analyze 3D and 4D image stacks generated by ScanImage microscopy software.
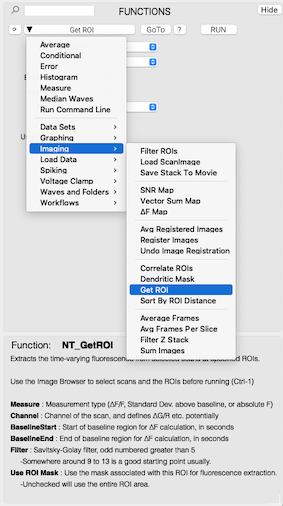
Analysis functions that are included in an imported package are automatically incorporated into the NeuroTools interface, and have access to all user-made data sets. For instance, below is an example of the SetScale function. By using a small amount of specialized syntax, the control layout--including resolving variable types, wave inputs, and menu items--are automatically generated from the written code, so the user doesn't need to manually define input controls.
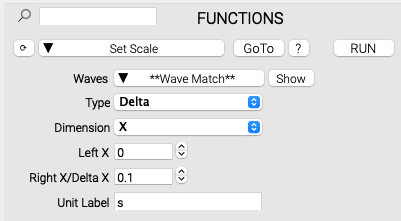
The user code is parsed by NeuroTools to determine how to neatly display its inputs in the GUI:
Function NT_SetScale(DS_Waves,menu_Type,menu_Dimension,Offset,Delta,ScaleStr) //TITLE=Set Scale //SUBMENU=Waves and Folders // Note={ // Sets the dimension scaling of the input waves. // } String DS_Waves // Data set input as a menu String menu_Type,menu_Dimension // Drop down menus Variable Offset,Delta // Numeric variable input String ScaleStr // String variable input // Define menu list items and control titles String menu_Dimension_List = "X;Y;Z;T;" String menu_Type_List = "Boundary;Delta;" String Offset_Title = "Left X" String Delta_Title = "Right X/Delta X" String ScaleStr_Title = "Unit Label" //Declare and fill the structure that holds the input data set waves STRUCT ds ds GetStruct(ds) Variable i For(i=0;i<ds.numWaves[%Waves];i+=1) // Declares each wave from the data set 'Waves' Wave w = ds.waves[i][%Waves] // Sets the scale of the input wave based on the menu selections strswitch(menu_Type) case "Boundary": strswitch(menu_Dimension) case "X": SetScale/I x,Offset,Delta,ScaleStr,w break case "Y": SetScale/I y,Offset,Delta,ScaleStr,w break case "Z": SetScale/I z,Offset,Delta,ScaleStr,w break case "T": SetScale/I t,Offset,Delta,ScaleStr,w break endswitch break case "Delta": strswitch(menu_Dimension) case "X": SetScale/P x,Offset,Delta,ScaleStr,w break case "Y": SetScale/P y,Offset,Delta,ScaleStr,w break case "Z": SetScale/P z,Offset,Delta,ScaleStr,w break case "T": SetScale/P t,Offset,Delta,ScaleStr,w break endswitch break endswitch EndFor End
Data Sets
Data sets can be defined using wave matching tools in the main interface or, for a more permanent solution, they can be directly defined using a data table. Data sets can be sent to a function as a whole, or in subsets, depending on how the user has organized the data set. For instance, consider a data set that has 500 waves--10 neural recording sessions, each with 5 repeats of a 10 voltage step stimulus. In a single click, you could send each set of 10 voltages to a function individually, one subset at a time, by organizing the data set using the wave grouping input "/B=10" :
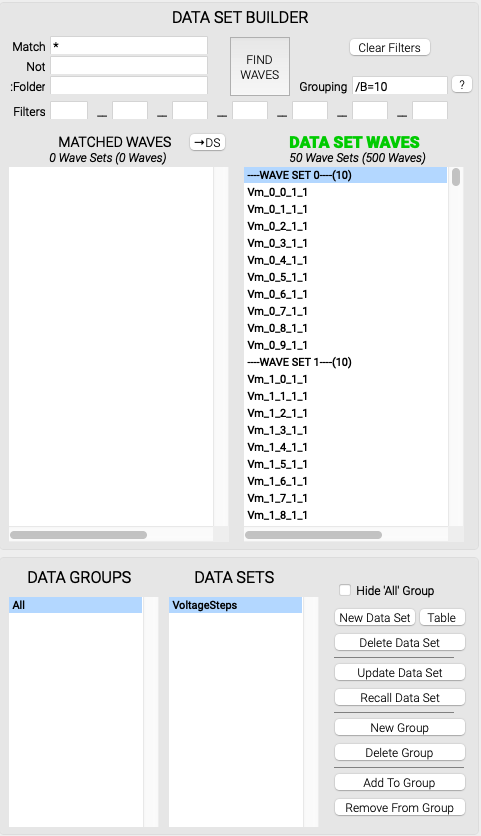
/B=n : Groups data in blocks of n
/S=n : Groups data with stride of n (every nth wave is in the same subset)
/WG=n : Groups data according to the wave name residing in the nth underscore position.
/A = n : Sorts data alphanumerically according to the wave name residing in the nth underscore position.
/WSI = n : Wave set index. Only includes the range nth wave, or range of waves, within each subset.
/WSN = n : Wave set number. Only includes the nth subset (i.e. wave set) or range of subsets.
These organizing tools are stackable, so /B=10/S=2 would first put the data in blocks of 10, and then organize each resulting subset by a stride of 2. This flexibility should allow a user to achieve nearly any arbitrary organizing principle.
Command line integration
NeuroTools has an integrated command line that has access to its data sets. This allows the user to perform single-use operations in batch. Using the above-mentioned VoltageSteps data set as an example:
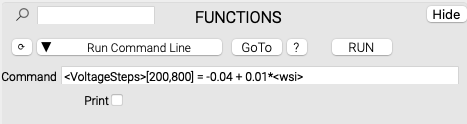
<VoltageSteps> references each wave in the data set called 'VoltageSteps', and executes the operation on it. <wsi> tells the command line to use the wave's index in the subset as the multiplier. This results in a step function from -0.04 incrementing by 0.01 for each wave in each subset.
Searchable Functions
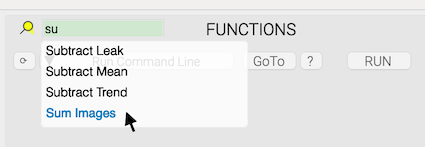
As more functions get added into NeuroTools with add-on packages, they can be slightly cumbersome to navigate to using the functions drop down menu. A live text search tool remedies this annoyance.
Installation
Unzip the file, open INSTALL_NEUROTOOLS.pxp in Igor. Installation and file distribution will be carried out automatically from there. Restart Igor, and the GUI can then be loaded from the NeuroTools menu.
Project Details
Current Project Release
| Release File: | NeuroTools+.zip (3.1 MB) |
| Version: | NeuroTools v3.14 |
| Version Date: | |
| Version Major: | 3 |
| Version Patch Level: | 14 |
| Release Link: | https://www.benmurphybaum.com/NeuroToolsStore.html |
| OS Compatibility: | Windows Mac-Intel |
| Release Notes: |
-Major improvements in ease of use, bug fixes, auto-installation. -Data tables are improved for more permanent storage of data sets. -Add-on analysis packages are simpler to add to NeuroTools. |

Forum

Support
Gallery
Igor Pro 10
Learn More
Igor XOP Toolkit
Learn More
Igor NIDAQ Tools MX
Learn More
